Biomolecular Information Science
Cooperation Course
Staff
Tomohiko Tsuge
- Position
- Associate Professor
- Office
- Uji Campus, Nucleic Acid Research Bldg (ICR), Room 5
- Phone
- 0774-38-3263
- Fax
- 0774-38-3259
- tsuge(at-mark)scl.kyoto-u.ac.jp
Mariko Kato
- Position
- Assistant Professor
- Office
- Uji Campus, Nucleic Acid Research Bldg (ICR), Room 5
- Phone
- 0774-38-3263
- Fax
- 0774-38-3259
- kato(at mark)scl.kyoto-u.ac.jp
Tomomi Fujii
- Position
- Assistant Professor
- Office
- Uji Campus, Main Bldg, Room N-236C
- Phone
- 0774-38-3258
- Fax
- 0774-38-3045
- fujii(at mark)scl.kyoto-u.ac.jp
Research
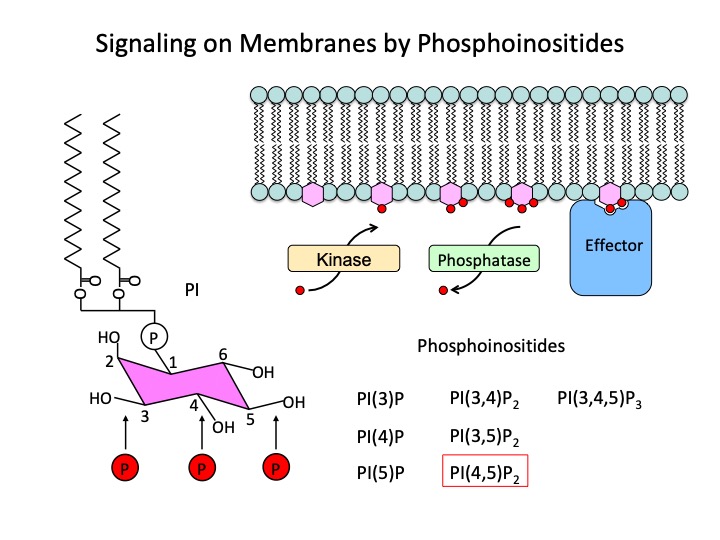
Control mechanism of plant cell morphogenesis by phospholipid signals
Phospholipids such as phosphoinositide are not only the main constituents of biological membranes in eukaryotic cells, but also function as signal molecules with intracellular position information by localizing to specific membrane regions. In elongating root hairs and pollen tubes, tip growth is positively controlled by the localization of PI (4,5) P2 on the cell membrane at the tip. It has also been reported that PI (4,5) P2 is localized in the upper and lower cell membranes of Arabidopsis root proliferating cells and is involved in the polar transport of auxin via clathrin-dependent endocytosis.
PIP5K, a PI (4,5) P2 producing enzyme, is encoded in 11 genes in Arabidopsis thaliana. In our laboratory, we select function-deficient mutants for each of these PIP5K genes and analyze the phenotypes of mutants containing these higher-order multiple mutants. Duplication of genetic function in plants often impedes genetic analysis. However, when it is involved in an event that is essential for cell proliferation and differentiation, such as PIP5K, it may be possible to obtain a phenotype in a specific phenomenon without having a fatal effect depending on the combination of multiple mutants. We hope to discover new functions of phospholipid signals that could not be captured by genetic analysis of animals and yeast.
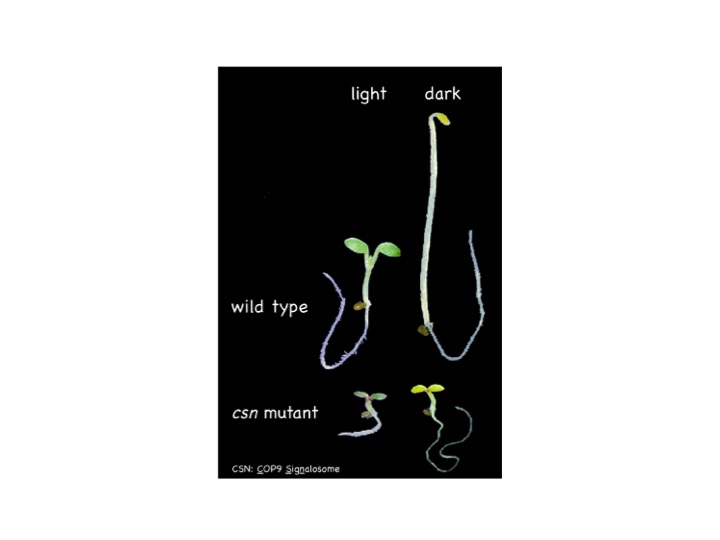
Plant plasticity control mechanism through mRNA metabolism
We are focusing on the light environment response of plants as a model for plastic shaping. We have shown that when animals and plants respond to the environment, COP9 signarosomes (CSNs) perform “quantitative control” that controls the degradation of transcription factors. CSN, on the other hand, binds to factors that control RNA splicing and polyadenylation. Therefore, we are elucidating the “qualitative regulation” of transcripts by CSN, focusing on alternative splicing and selective polyadenylation of RNA. Specifically, we are practicing a research style that combines mutants and transformed plants and makes full use of molecular biological techniques such as anatomy and biochemistry. We are unraveling the dynamic and highly universal molecular mechanism in the shaping of living things.
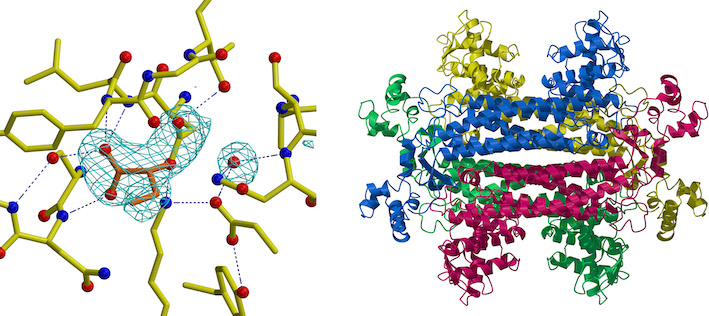
Elucidation of environmental adaptation strategies based on protein molecular structure
We mainly determine the three-dimensional structure of protein molecules using the method of X-ray crystal structure analysis, and conduct structural biology research on the relationship between the structure and its functions and physical properties. Our main research themes are to elucidate the substrate recognition mode and catalytic reaction mechanism of enzymes, and to elucidate the environmental adaptation strategy of microbial-derived proteins that grow in extreme environments of high or low temperatures.